Turbocharging the AAA+ ATPase Motor: Zairong Zhang’s Lab Identifies a Power-Boosting Module Essential for Ubiquitin-Mediated Protein Degradation
Date:2025-11-25
Many macromolecular assemblies—such as membrane protein–lipid bilayers, protein–nucleic acid complexes, and large protein assemblies—are structurally robust and tightly organized. To deliver their protein components to the proteasome for degradation, cells must first extract these substrates intact from their native multi-component environments, a process that requires mechanical force generated by dedicated molecular machines. The AAA+ ATPase p97/VCP plays a central role in this extraction step. Over the past two decades, p97/VCP in complex with its cofactors UFD1L–NPLOC4 (UN) has been recognized as the core “segregase” capable of unfolding highly polyubiquitinated substrates. However, whether p97/VCP–UN alone provides sufficient mechanical power to pull substrates out of hydrophobic lipid bilayers or densely packed protein–nucleic acid assemblies has remained unclear.
A recent study from Dr. Zairong Zhang’s group at the Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, published in Nature Communications, uncovers the key mechanism by which p97/VCP gains the power necessary to carry out these demanding tasks. The authors identify a previously unknown ubiquitin-binding helix (UBH) present in many UBX/UBXL family cofactors—such as FAF2—across yeast, plants, and metazoans. Together with the adjacent UBX domain, UBH forms a UBH–UBX module that doubles the mechanical output of p97/VCP, thereby enhancing its substrate-unfolding ability and enabling efficient extraction of membrane-embedded proteins.
To search for factors that regulate p97/VCP ATPase activity, the Zhang lab used tandem affinity purification and identified FAF2 as an adaptor that physically associates with both p97/VCP and its ubiquitinated membrane substrates. FAF2 is distributed on multiple organellar membranes—including the ER, mitochondria, peroxisomes, and lipid droplets. Traditionally, FAF2's UBX domain was thought to primarily recruit p97/VCP to membrane surfaces, while the UBA domain was assumed to recognize ubiquitinated substrates; the long helical domain between them had no clearly defined function.
The Zhang team showed that FAF2 is indispensable for ER-associated degradation of misfolded proteins. Surprisingly, deleting the UBA domain had little effect on degradation, whereas removing the long helical region caused a dramatic defect. Further experiments revealed that this helical region is in fact a novel ubiquitin-binding element—UBH—that preferentially recognizes K48-linked polyubiquitin chains. Acting together with the adjacent UBX domain, UBH cooperates with UN to enhance ubiquitin-chain engagement and ensure efficient substrate delivery to p97/VCP. Importantly, the UBH–UBX module is not unique to FAF2; it is broadly conserved in other p97 adaptors, including FAF1.
Through reconstitution experiments, the team uncovered an important but previously overlooked principle: although p97/VCP–UN can bind ubiquitinated membrane proteins, it is not able to extract them from the lipid bilayer through its own power. When FAF2—or specifically its UBH–UBX module—is supplied, ATP hydrolysis by p97/VCP increases markedly, substrate unfolding is enhanced, and membrane proteins can finally be removed from the bilayer. In essence, UBH–UBX acts as a “power amplifier” for the p97 motor, enabling it to handle difficult substrates that would otherwise resist extraction.
Intriguingly, UBH homologs are present in UBX-family proteins across plants, yeast, insects, and mammals. Cross-species analyses confirm that these helices bind K48-linked ubiquitin chains, suggesting that this mechanism is a deeply conserved evolutionary solution for challenging protein extraction tasks. Moreover, UBH–UBX function extends beyond ERAD: UBH mutations impair degradation of the mitochondrial outer membrane protein MCL1, highlighting its essential role in mitochondrial-associated degradation (MAD). Loss of UBH also promotes excessive pexophagy and reduces peroxisome abundance, underscoring its importance in maintaining peroxisome homeostasis.
This study is the first to reveal that the long helical domains in UBX proteins such as FAF2 and FAF1 are actually novel ubiquitin-recognition modules forming a p97/VCP “power-boosting unit.” UBH–UBX grants p97/VCP the mechanical force necessary to extract ubiquitinated proteins from large biological assemblies. These findings fill a major gap in our understanding of p97/VCP function and provide a unifying explanation for its roles across ERAD, MAD, peroxisome quality control, chromatin-associated degradation, and other pathways. Given the strong connection between p97/VCP dysfunction and neurodegeneration, myopathies, and viral infection, the UBH–UBX module represents an attractive new target for developing pathway-selective modulators to treat disorders involving proteostasis imbalance.
This work was supported by the National Natural Science Foundation of China, the Chinese Academy of Sciences, the Shanghai Science and Technology Commission, and benefited from extensive support by the Zhang Yaoyang group and Pan Lifeng group at SIOC.
Original article: https://www.nature.com/articles/s41467-025-65166-4
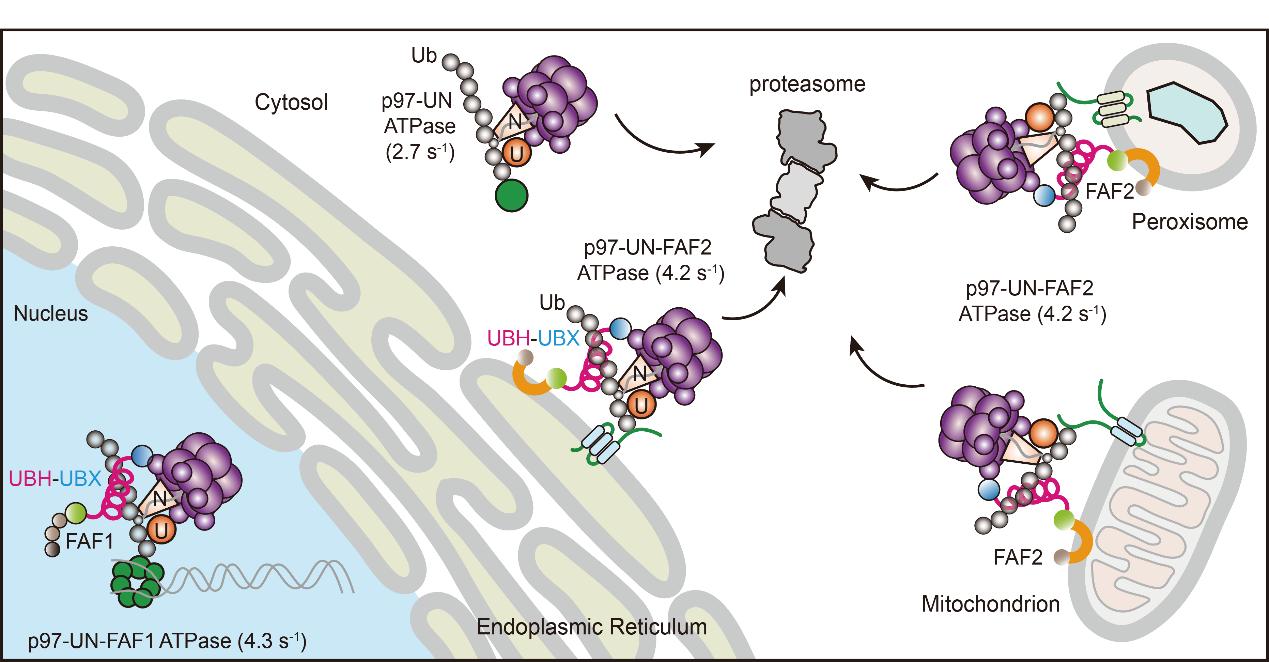
Schematic model of how FAF2 and FAF1 regulate organelle homeostasis and replisome disassembly
附件下载:
