The Ma lab’s research focuses on the intersection of animal behaviors and molecular neurobiology, utilizing advanced single cell technologies. Our primary interest lies in understanding how heterogenous gene expression in clock neurons leads to different rhythmic behaviors such as locomotor activity and sleep.
The circadian clock is an evolutionarily conserved and adaptive feature found in many eukaryotic organisms. It confers fitness by a daily adjustment of its not exactly 24 hours core molecular pacemaker to the environmental fluctuations such as light/dark cycles, temperature, or nutrition. The molecular pacemaker is most notable in a restricted region named suprachiasmatic nucleus (SCN) in mammals. There is a comparably important circadian region in the fly brain, the ca. 150 Drosophila clock neurons. The pacemaker drives a broad sweep of rhythmic gene expression, which plays a central role in animal physiology, metabolism and behaviors including locomotor activity and sleep.
Using a cutting-edge single cell RNA sequencing method, we have previously demonstrated the remarkable heterogeneity in gene expression within the fly clock neurons. Comparative analysis between clock and dopaminergic neurons has revealed that neural connectivity molecules, such as GPCRs and cell surface molecules best identify the transcriptomic cell types in the fly brain. We are working to understand the mechanism through which cell type specific gene expression regulates animal behaviors and how core clock proteins govern rhythmic gene expression in different cell types.
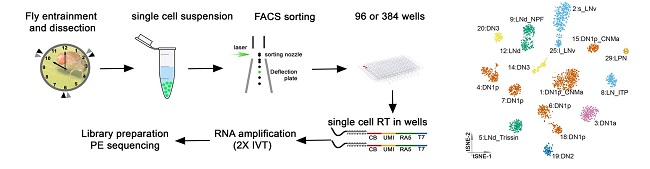
Single-cell RNA sequencing reveals the spatiotemporal heterogeneity of rhythmic neuronal gene expression.
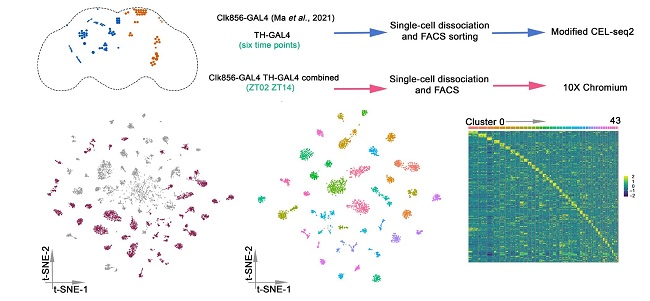
Comparative study of clock neurons and dopamine neurons in the adult fruit fly brain at the single-cell level.
